Fig. 1
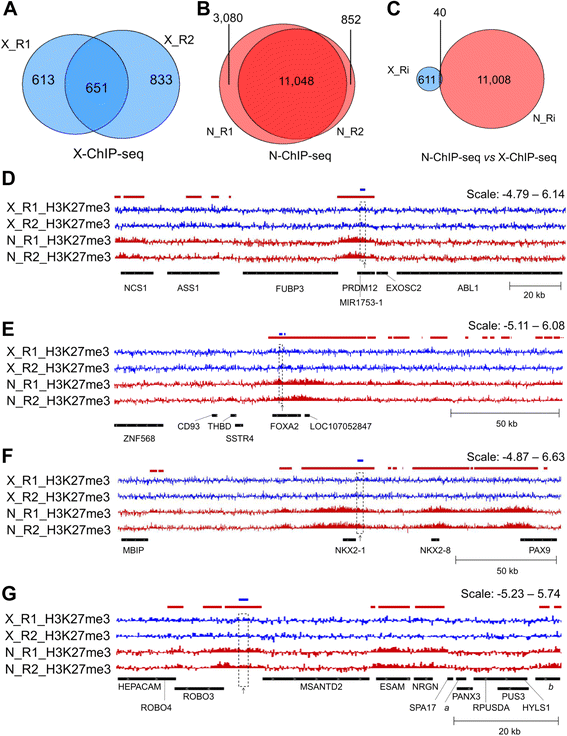
N-ChIP-seq and X-ChIP-seq results. a-c Venn diagrams representing the overlap determined by intersectBed between H3K27me3 broad peak regions detected by epic. a X-ChIP-seq replicates. b N-ChIP-seq replicates. c Intersection between X-ChIP-seq and N-ChIP-seq peaks (only common peaks for each condition were considered). d-g Visualization with IGV of H3K27me3 enrichment normalized to input [log2(IP/input)]. X-ChIP-seq muscle tracks are shown in blue and N-ChIP-seq muscle tracks in red. The colored boxes above the tracks represent broad peaks detected by epic in either X-ChIP-seq (blue) or N-ChIP muscle (red) replicates. The dashed boxes across the tracks represent the localization of a common peak (between methods). The black arrows under the tracks represent the position of qPCR primers used for Fig. 2A. d Common peak 1 (chr17: 6,422,600–6,424,599); (e) common peak 2 (chr3: 3,359,200–3,360,599); (f) Common peak 3 (chr5: 36,681,000–36,683,799); (g) common peak 4 (chr24:262,600–264,199), a: LOC107055042, b: LOC101748751