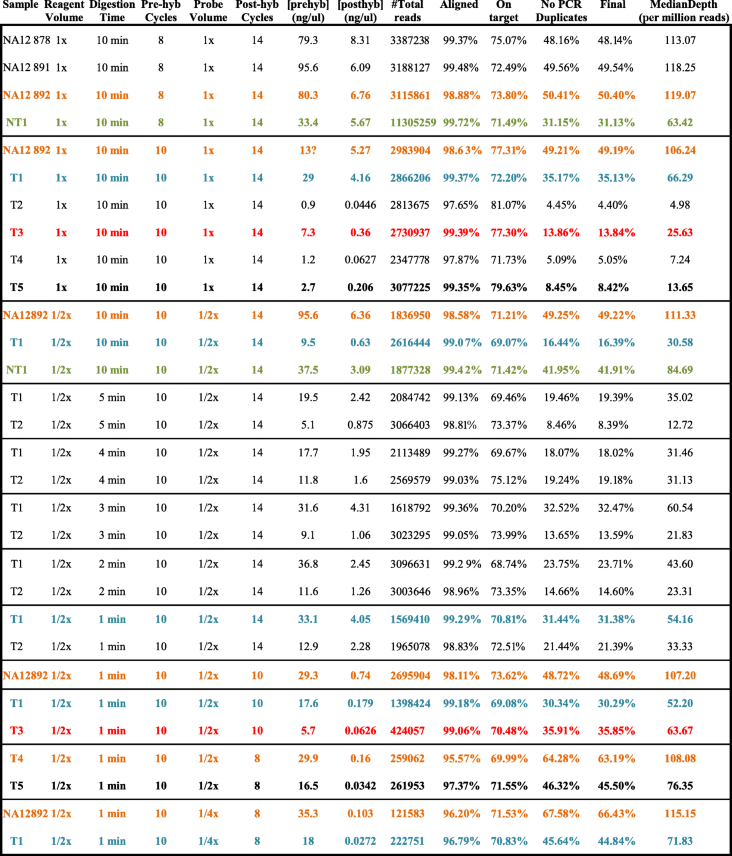
- From left to right: sample name, reagent volume (1× = standard, 1/2× = half standard), digestion time, number of cycles in pre-hyb PCR, volume of probe (1× = standard, 1/2× = half standard, 1/4× = quarter standard), number of cycles in post-hyb PCR, pre-hyb library concentration, post-hyb library concentration, number of sequences obtained, percentage of sequences aligned to human reference genome, percentage of sequences on target, percentage of sequences after PCR duplicates removal, percentage of sequences after low-quality-mapped-reads removal, median of the depth in target regions per million reads (normalized value for comparison). In orange, results for sample NA12892 highlighting that no negative impact was noticeable after modifications of the protocol in high-quality DNA samples. In green, improved results obtained for a high-quality FFPE sample (NT1) with some of the modification. In blue, results for sample T1 with notable improvement obtained after modifications. In red and bold, improved results for two very low-quality FFPE samples (T3 and T5) after modifications in the protocol
